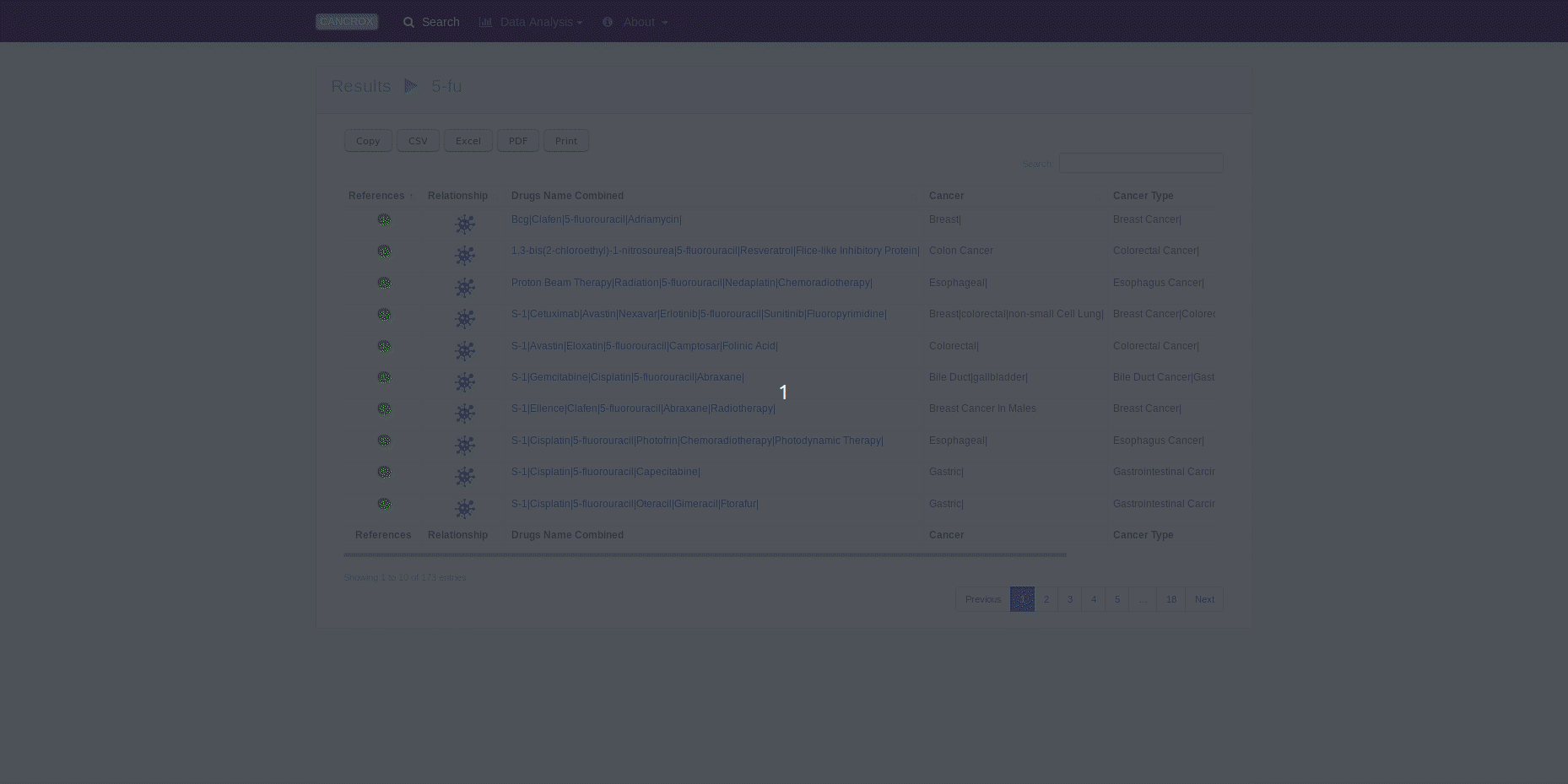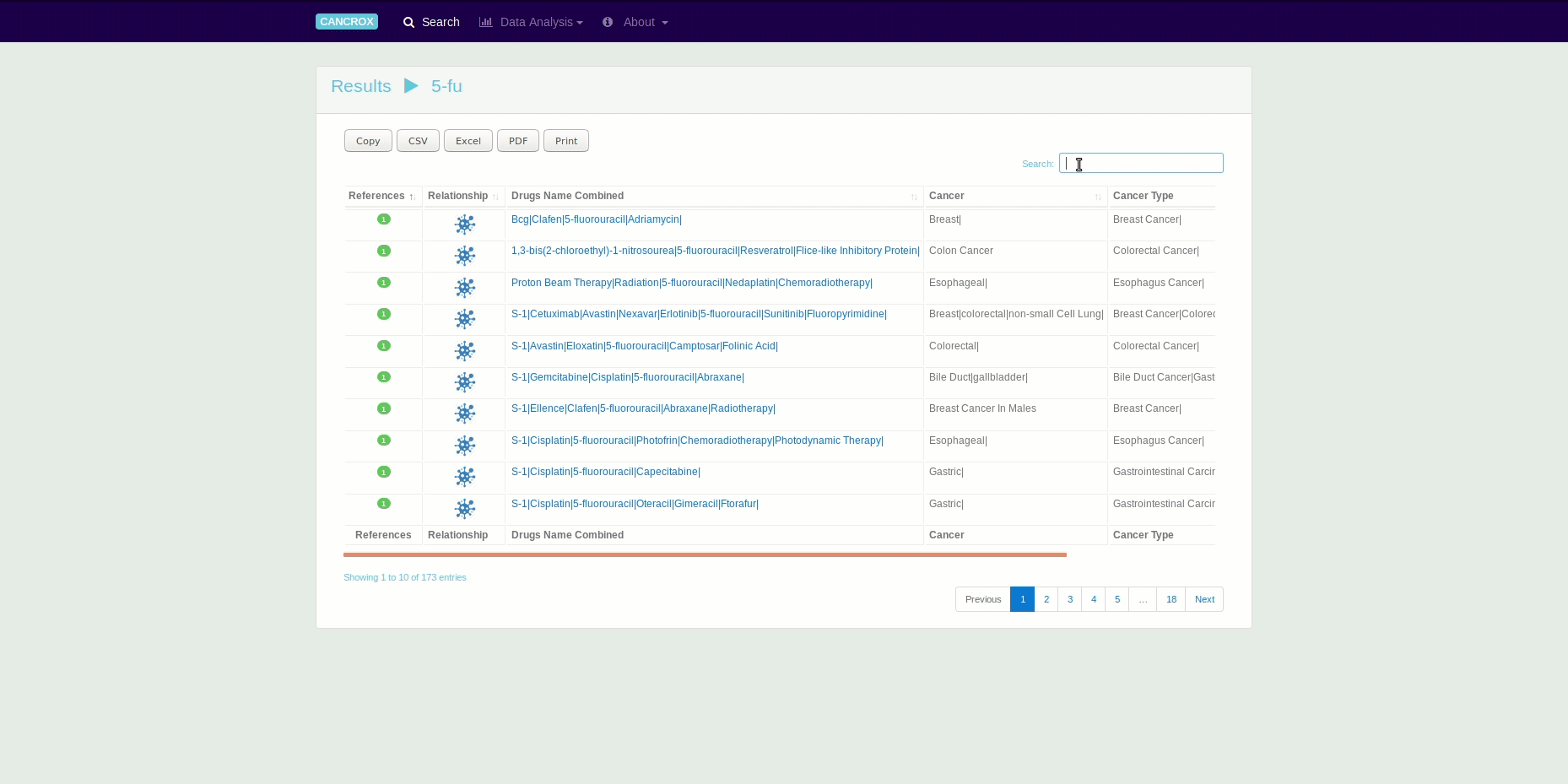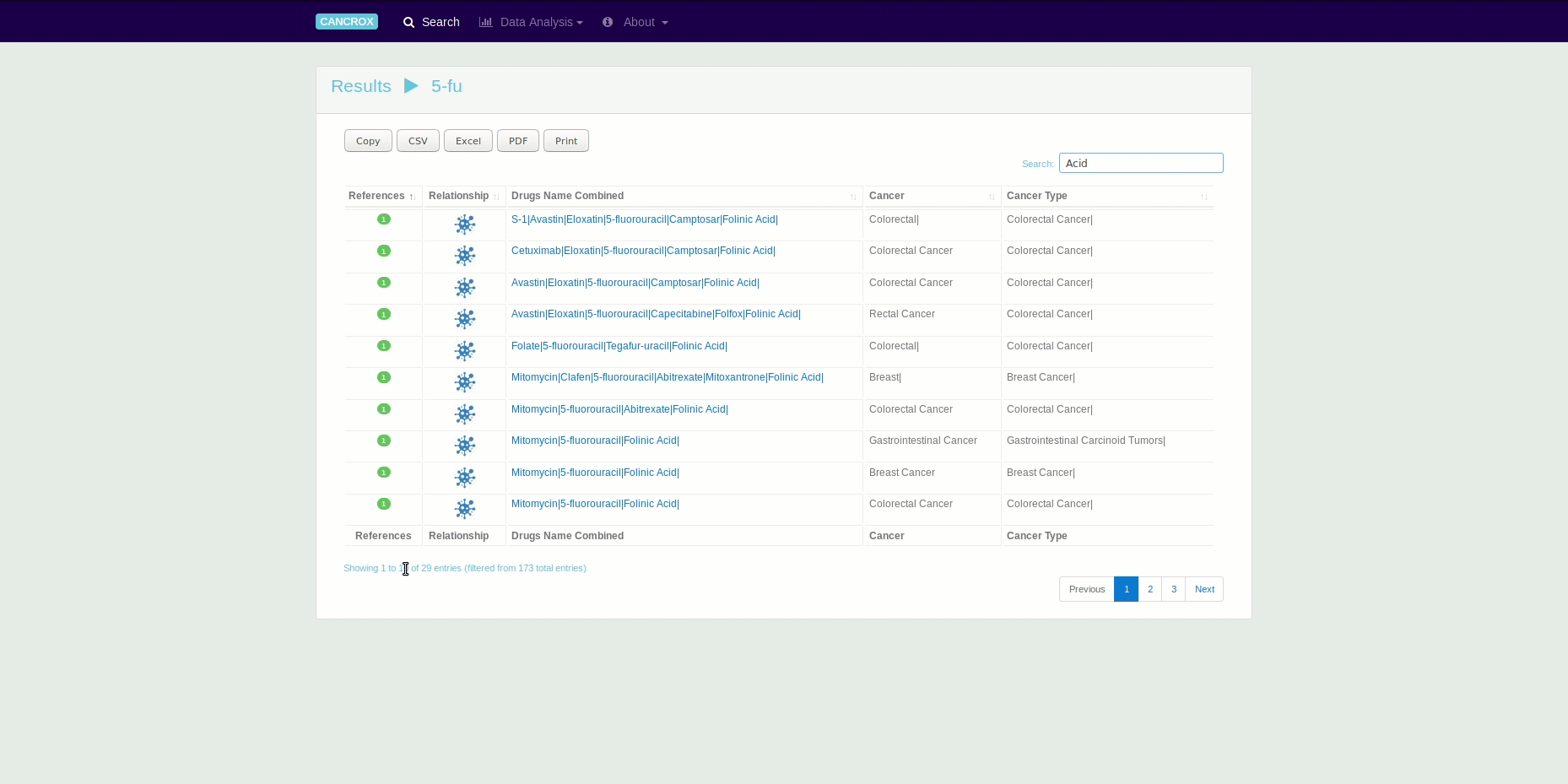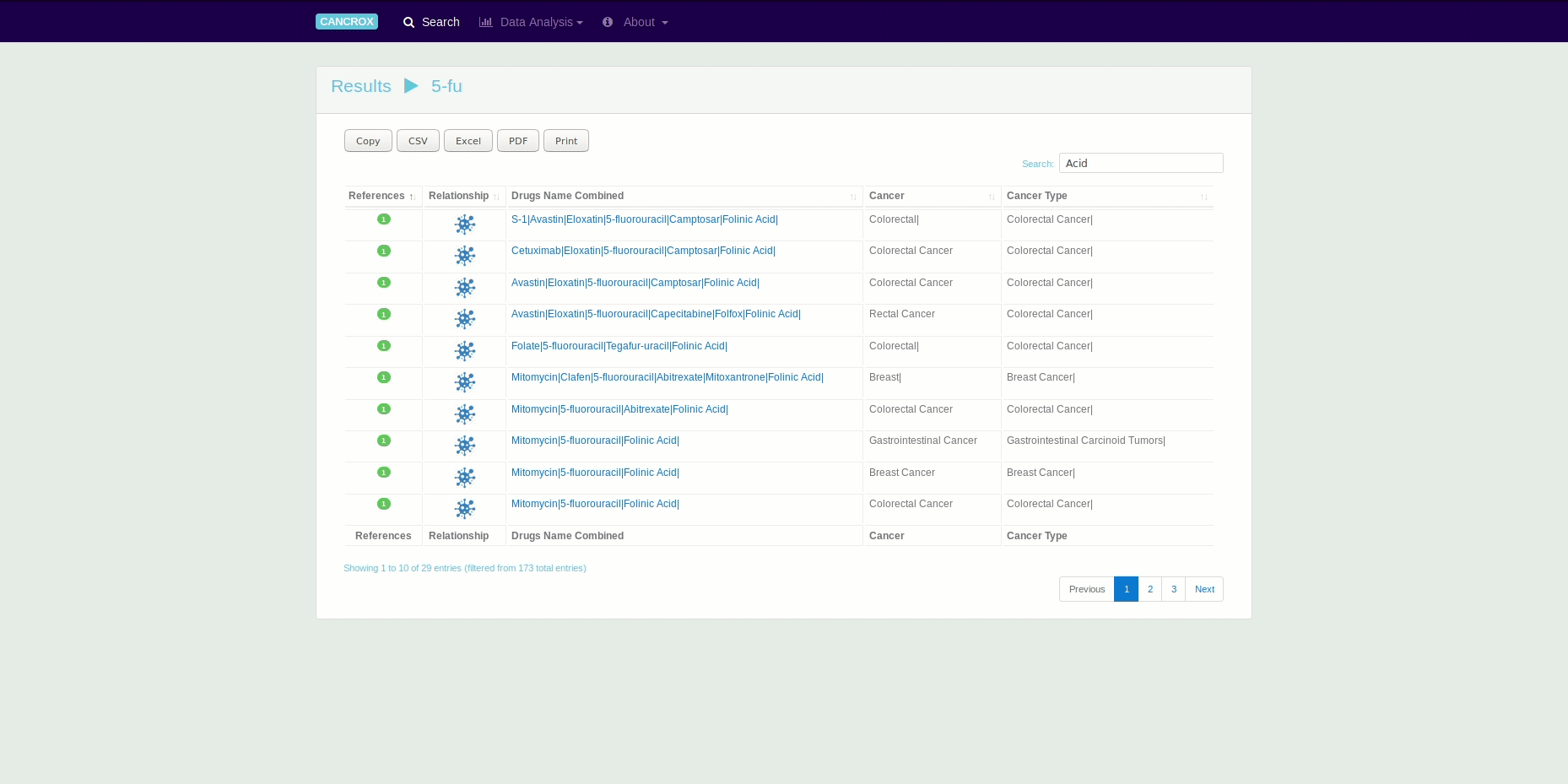
The CANCROX tool is able to perform TM and NLP analysis in thousands and even millions of scientific articles, constructing a reference database of similar genes between humans and dogs associated with cancer, hereinafter referred to as candidate genes. In addition to candidate genes, the types of cancer associated with these genes, therapies, and drugs and their different combinations are identified and stored in this database.
In the table below you can download CSV files separated by semicolons.
Main Files
| Drugs and Cancer |
This file shows the combination of drugs and therapies associated with cancers. The 'drugs_title_conclusion_id' field lists the drug code. The list of codes and names is available in this file. |
| Candidates Genes |
This file presents the listing of 477 similar candidate genes between humans and dogs and associated with various types of cancer. The original listing of genes associated with cancers can be obtained from the Sanger Institute. |
Auxiliary Files
| Code (Ids) of cancer types |
This file presents the relationship between cancer type codes (IDs) and their nomenclature. |
| Code (Ids) of drugs and therapies |
This file presents the relationship between drug codes and therapies and their respective nomenclatures. It establishes a relationship between the codes of the bases: Drugbank, Pubchem and ChemSpider. |
Classifier: Test and Train data
| Classifier Dataset |
This file provides test and training files used to implement the model produced by the Random Forest algorithm. |
NER: Test and Train data
| Ner Datasets |
This file provides the complete set of abstracts annotated in the Apache OpenNLP library standard. The developer can create a script to prepare the test and training files. |
It is possible to obtain data in different formats using the search engines and by exporting the data directly from the tables. See an animated gif presented this feature.